Numpy and Simulations
In [1]:
%load_ext interactive_system_magic
decay.py
import numpy as np
def initialise_state(num_steps, initial_value):
"""
Args:
num_steps (int): the number of iterations to run the simulation for
initial_value (float): the value to start decaying from
Returns:
np.array: a container for the system state with the initial state present
"""
state = np.zeros(num_steps)
state[0] = initial_value
return state
def update_state(old_state):
"""
Args:
old_state (float): the previous value
Returns:
float: the value, decayed
"""
DECAY_RATE = 0.9
new_state = old_state * DECAY_RATE
return new_state
def run_simulation(num_steps, initial_value):
"""
Runs the simulation using the rules defined in ``update_state``.
Args:
num_steps (int): the number of iterations to run the simulation for
initial_value (float): the value to start decaying from
Returns:
np.array: the overall state for the system
"""
state = initialise_state(num_steps, initial_value)
for t in range(1, num_steps):
state[t] = update_state(state[t-1])
return state
INITIAL_VALUE = 100
NUMBER_OF_STEPS = 100 # Set this to 100
value_history = run_simulation(NUMBER_OF_STEPS, INITIAL_VALUE)
np.savez_compressed("decay", state=value_history)
$
python decay.py
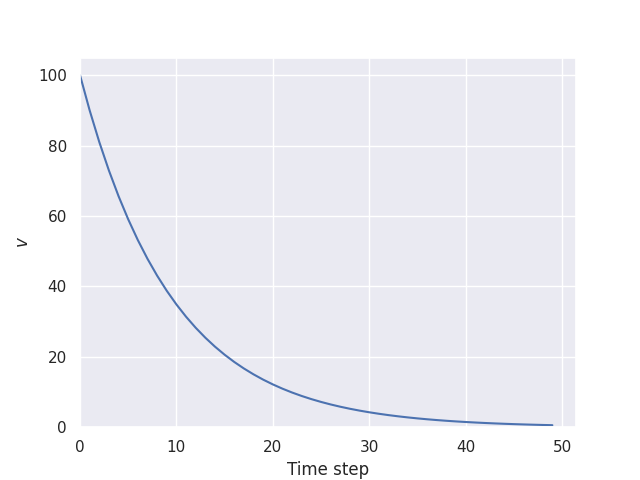
We can then load the data and plot it. We can do this either in a Jupyter Notebook cell or in another python script:
decay_plot.py
import numpy as np
import seaborn as sns
sns.set_theme()
with np.load("decay.npz") as f:
value_history = f["state"]
g = sns.relplot(data=value_history, kind="line").set(
xlim=(0,None),
ylim=(0,None),
xlabel="Time step",
ylabel="v"
)
g.savefig("decay.png") # Save the graph with this line
$
python decay_plot.py
This has created a file called decay.png: